The application of molecularly targeted cancer therapies has rapidly expanded in the past 2 decades. The identification of genetic mutations in tumors has led to a growing list of therapies developed to specifically target those alterations. Indeed, molecularly targeted therapies—along with predictive biomarker testing and site-specific chemotherapy—have significantly improved the outcomes of patients with certain tumor types. Improved patient progression-free survival (PFS) and overall survival have been reinforced by the rapid evolution of genomic testing. Specifically, Sanger sequencing of a single gene has given way to broad-based genetic sequencing, or next-generation sequencing (NGS), to sequence millions of small fragments of DNA simultaneously.
The hope was that the widespread availability and adoption of NGS would revolutionize guided cancer treatment. This future is rapidly becoming reality as clinicians have greater access to the technology and are increasingly sequencing patients’ tumor genomes. Using this information, they are able to identify mutations that may influence patients’ cancer genesis and progression, before selecting personalized treatments matched to these mutations.
Notably, improvement in outcomes has been achieved with agents matched to a molecular alteration in specific tumor types. The discovery that similar mutations can occur in tumors derived from different tissue types1 has led to the consideration of common therapies across tumors of distinct origin that harbor the same mutation. This shift toward histology-agnostic molecularly targeted treatment has led to the concept of basket trials. These trials are designed to test whether a specific molecular alteration in a patient with cancer will respond to a matched agent. As part of a basket trial, a common mutation across a range of tumor types is treated using similar therapy.
Results from the first basket trials have led to a growing realization that a single tumor mutation may not be driving the growth of a particular tumor type because there is an abundance of passenger mutations also present whose roles are not always clearly understood. In addition, advances in—and broad access to—NGS has rapidly outpaced therapeutic development timelines. These factors suggest that not all mutations identified by NGS are clinically actionable, and may present a challenge for practicing oncologists trying to choose the best cancer treatment option for their patients. Identification of ≥1 tumor mutations may also lead to unrealistic patient expectations regarding the availability of therapies and their prognosis.
To make an appropriate treatment selection, the extensive genetic information extrapolated from NGS requires context, including the site of origin of the cancer. Knowledge of tumor type and primary site of origin of the cancer are proving to be critical for oncologists making treatment decisions. Without this information, oncologists would not be able to determine the potential benefits of site-specific and molecularly targeted therapies, which may lead to less than optimal responses, and expose patients to toxicities and unnecessary costs.
Epidemiologic studies have shown that the site of origin of the tumor is unknown or uncertain in approximately 1 of 6 patients with newly diagnosed metastatic cancers.2-5 Situations where identification of the site of origin is particularly challenging include poorly differentiated or undifferentiated tumors, tumors with an atypical clinical presentation, or tumors with limited biopsy tissue for immunophenotypic analysis.
Difficult-to-diagnose tumors make therapeutic decision-making challenging, but the use of molecular profiling assays can play an important role in establishing a definitive diagnosis, and ensuring that a patient is given the most appropriate site-specific chemotherapy and targeted therapies.
Results from Recently Reported Basket Trials
Clinical trials have demonstrated that genomic information is useful for guiding the development of novel targeted agents, as well as treatment decisions. Basket trials have arisen as a proposed alternate approach to clinical trials by more efficiently testing cancer therapies for new uses. However, the results from the first of these studies question the role of specific alterations across multiple tumor types.
Proof-of-Concept Basket Trial: No Improvement
in PFS
In the SHIVA basket trial, Le Tourneau and colleagues conducted a histology-agnostic study using molecular information to guide therapy in patients with any type of metastatic solid tumor that is refractory to standard treatment.6 Patients underwent molecular profiling of a metastatic tumor, and were eligible for inclusion in the study if they had an aberration in the hormone receptor, phosphoinositide 3-kinase/protein kinase B (AKT)/mechanistic target of rapamycin, or RAF/MEK pathways. Patients were randomized to receive treatment with a molecularly targeted agent matched to the alteration, or treatment at their physician’s choice.
The investigators found that median PFS was not improved in the experimental group compared with the control group (2.3 months vs 2.0 months; P = .41). The study authors concluded that, in heavily pretreated patients with cancer, using molecularly targeted agents outside the realm of their indications does not improve PFS compared with treatment selected at a physician’s choice.
Altogether, these data suggest that although molecularly targeted agents are linked to antitumor activity in patients with tumors that have matching molecular alterations, off-label use of these agents based on matching molecular alterations alone may not be warranted.
CUSTOM Trial Showed Some Promising Results, but Highlighted Challenges in Basket Trial Design
The CUSTOM (molecular profiling and targeted therapy for advanced thoracic malignancies) basket trial focused on identifying molecular biomarkers and determining their frequency and clinical relevance in patients with advanced thoracic malignancies.7 The investigators evaluated the efficacy of multiple targeted therapies in specific molecular subsets of patients with advanced non–small-cell lung cancer (NSCLC), small-cell lung cancer, and thymic malignancies.
A total of 647 patients met the general eligibility criteria of the trial, and underwent molecular profiling. Although the study design was appealing because the investigators assessed the use of 5 drugs (erlotinib, selumetinib, MK-2206, lapatinib, and sunitinib) matched against specific mutational status, only 43 of the patients received treatment, and were evaluated for response and survival. A 60% response rate was seen in patients with epidermal growth factor receptor (EGFR)-mutated NSCLC who were treated with erlotinib (n = 15).
Mixed Results in Nonmelanoma BRAF V600-Mutated Cancers
In another basket trial, investigators enrolled 122 patients with nonmelanoma cancers and BRAF V600—a mutation that occurs in approximately 50% of cutaneous melanomas.8 Patients received treatment with vemurafenib or vemurafenib plus cetuximab. BRAF V600 mutations were targetable in some, but not all, of the nonmelanoma cancers tested, the investigators observed. Vemurafenib activity was seen predominantly in patients with BRAF V600–positive NSCLC, Langerhans cell histiocytosis, and Erdheim-Chester disease. In a subset of patients with colorectal cancer (n = 27) who received vemurafenib monotherapy, there were no responses, and the median PFS and overall survival were 4.5 months and 9.3 months, respectively. None of the patients with multiple myeloma had a response. Altogether, the data did not support the assumption that one can extrapolate the effectiveness of a targeted therapy to all cancer types with the same mutation. “What we’ve learned is that the driver mutation matters, but the tissue of origin is also important,” explained Baselga and colleagues in their study.9
In addition to basket trials, other studies have raised questions about the role alterations, in particular tumor types, play across multiple tumors. In particular, the success of BRAF inhibition in melanoma served as a catalyst for investigating vemurafenib in patients with BRAF-mutant colorectal cancer. In that study, the investigators purported that the same inhibitors that had worked so well in patients with melanoma would translate to activity in the 5% to 8% of patients with colorectal cancers that harbor the same BRAF V600E mutation.10 A pilot study of vemurafenib in 21 patients with BRAF V600–mutant metastatic colorectal cancer proved disappointing, however, with no complete responses, only 1 partial response, and 7 cases of stable disease. The median PFS was 2.1 months, and median overall survival was 7.7 months. The robust clinical response of vemurafenib observed in melanoma was absent in BRAF V600–positive metastatic colorectal cancer.
Implications of Unclear Diagnoses in Patients with Metastatic Cancer
Diagnostic ambiguity in cancer is relatively underappreciated. A 2014 study from The University of Texas MD Anderson Cancer Center, Houston, TX, showed that in approximately 1 of 4 cases, diagnostic errors were identified during expert pathology subspecialist review.11 In a separate meta-analysis examining the performance of immunohistochemistry (IHC) in determining tissue of origin in metastatic cancers, IHC was inaccurate in 1 of 3 cases, particularly in poorly differentiated lesions.12,13
Of the 600,000 patients presenting with metastatic cancer each year, approximately 100,000 have some degree of diagnostic uncertainty, which translates to 1 of every 6 patients with metastatic cancer.2,3 This includes a variety of clinical situations which present with diagnostic ambiguity, including cancers of unknown primary (CUP), challenging differential diagnoses dealing with the main type of cancer (eg, pancreaticobiliary vs intestinal vs CRC) or subtype (eg, neuroendorine tumors subtyping: carcinoid or islet cell), lack of resolution between new primary versus recurrent cancers, tumors with an atypical presentation, and those with limited biopsy tissue.
A diagnosis using IHC can be especially difficult in certain scenarios; high-grade tumors, atypical metastasis, or poorly differentiated tumors can all present challenges in identifying the origin of the tumor. Even after multiple cycles of testing, many of these patients have an uncertain diagnosis. This uncertainty may lead clinicians to obtain a second diagnostic opinion, but doing so leads to a change in the treatment plan in only approximately 10% of cases.14 Furthermore, IHC staining may deplete tissue that may be essential for downstream predictive biomarker analysis.
Evidence from the aforementioned basket trials, and other data collected to date, suggest that strategies where patients with common mutations in disparate tumor types are treated similarly are not optimal. The implication is that patients without a single unambiguous diagnosis may be exposed to the side effects and costs of the treatment, potentially without tumor-suppressing benefits.
The use of targeted therapies in patients with unclear diagnosis is challenging because it is difficult to know whether mutations detected and acted upon are actually driving the cancer. These approaches can also delay getting the right therapy to the patient, and delays in appropriate treatment can affect prognosis. A definitive diagnosis, therefore, is critical for use of targeted therapies.
Improving Diagnostic Certainty: The Role of Molecular Assays in Challenging Cases
Patients with cancers of unknown or uncertain primary sites of origin may benefit from the use of molecular-based assays, by improving the accuracy and efficiency of diagnostic classification, and enabling the effective use of personalized treatment recommendations.
A gene expression based assay—CancerTYPE ID—has been developed to aid in resolving challenging diagnoses. CancerTYPE ID measures the differential expression of 92 genes (87 tumor-associated genes, and 5 reference genes) in formalin-fixed, paraffin-embedded (FFPE) tumor samples to provide a molecular classification of tumor type and subtype, and can distinguish between 28 main tumor types and 50 subtypes that represent >95% of all solid tumors by incidence. The 92 gene set was discovered through a genome-wide, data-driven approach.
CancerTYPE ID requires limited specimens—approximately 300 nonnecrotic cancer cells that can be collected from surgical resection, excisional biopsy, core needle biopsy, fine needle aspiration, cell block (pleural effusion, ascites), or bone marrow biopsy. Tumor cells are enriched by microdissection, and total ribonucleic acid (RNA) is isolated from FFPE specimens. RNA is then analyzed for the expression of the 92 genes using real-time, reverse transcriptase–polymerase chain reaction.
A computational algorithm compares the gene expression profile to a reference database with the gene expression profiles of >2000 tumors. The algorithm determines the most similar relative gene expression patterns, and predicts tumor type and subtypes. The algorithm assigns a probability for each classification result—which is a measure of the confidence level for the result—and rules out most tumor types in the database with >95% confidence. Test reports are typically available within 5 business days, and include a prediction of tumor type and histologic subtype.
CancerTYPE ID is a covered benefit for Medicare Part B patients based on a review of evidence supporting its analytical validity, clinical validity, and clinical utility.
Performance of the CancerTYPE ID Molecular Classifier
More than 23,000 patients have been analyzed by CancerTYPE ID, and the test has been ordered by >6000 oncologists and 5000 pathologists. The clinical development program has investigated >5200 cancer cases.
In a prospectively defined, blinded study of clinically challenging samples (ie, metastatic tumors, poorly differentiated and undifferentiated primary cancers, limited biopsy specimens), Sarah E. Kerr, MD, and colleagues demonstrated the sensitivity of a CancerTYPE ID of 87% for main tumor types, with no difference in sensitivity based on disease type (metastatic or primary), histologic grade, or in patients with a limited biopsy specimen, and a specificity of 98% to >99%.15
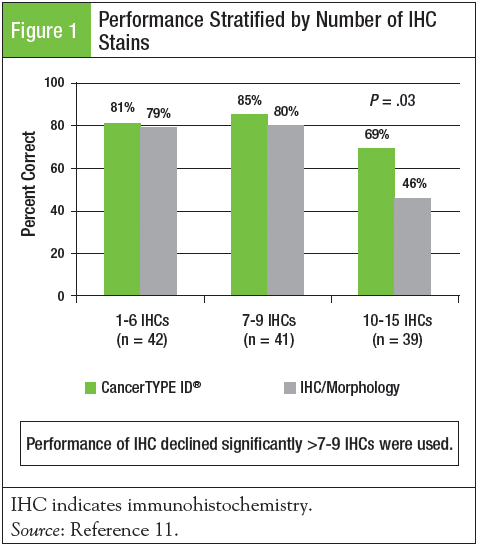
In a comparison to standard of care (IHC and expert pathology review), Lawrence M. Weiss, MD, FCAP, and colleagues conducted a blinded comparator study of patients typically referred for a diagnostic second opinion.11 A total of 122 patients with metastatic, poorly differentiated to undifferentiated tumors were submitted for evaluation by IHC and CancerTYPE ID with pathologists blinded to the CancerTYPE ID result. The mean number of IHC stains required to establish a diagnosis was 7.9. The overall accuracy of CancerTYPE ID was significantly superior to IHC/morphology (79% vs 69%; P = .03). The performance of CancerTYPE ID was superior or equivalent to IHC in all tumor types examined. The performance of IHC declined substantially when >7 to 9 IHC stains were used, suggesting that if classification of a single tumor type is not established by this point, CancerTYPE ID should be considered ahead of further staining (Figure 1).
Improving Treatment Outcomes in Patients
with CUP
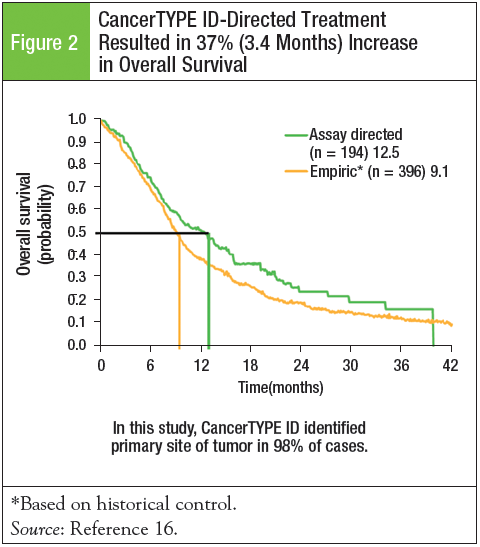
Only 1 prospective study completed in patients diagnosed with CUP has used a molecular classifier to predict the tumor type, and examined the efficacy of site-specific therapy based on the molecular assay results. John Hainsworth, MD, F. Anthony Greco, and colleagues evaluated the impact of CancerTYPE ID on patient survival in 252 patients with a diagnosis of CUP.16 Standard, first-line treatment regimens were applied based on the molecular results. CancerTYPE ID identified the primary tumor site in 98% (n = 247) of cases, and 26 different tumor types were predicted. The most commonly predicted tumor types—biliary tract, urothelium, colorectal, and NSCLC—accounted for 55% of all patients. In addition, 4 potentially curable germ cell types were predicted by the molecular assay. Among the 233 candidates for treatment, 194 received CancerTYPE ID–directed treatment. In these 194 patients, overall survival improved by 37%, and median survival was 12.5 months and 9.1 months in patients using assay-directed therapy versus the historical controls using empiric therapy, respectively (Figure 2).16
In a second outcomes study, the use of CancerTYPE ID was examined as an aid in the selection of optimal therapy for patients with CUP whose cancer was predicted to be of colorectal origin by CancerTYPE ID.17 Of the 42 patients for whom information on therapy and outcomes was provided, patients who received site-specific first-line treatment had a 50% response rate and 8.5-month median PFS. Patients who received empirical first-line therapy for CUP had a 17% response rate and 6-month median PFS.
Median survival in patients receiving site-directed therapy was 27 months, which was similar to survival in patients with known metastatic colorectal cancer, and substantially better than the median survival of 8 to 11 months in patients treated with empirical regimens for CUP.
Real-World Experience with CancerTYPE ID
Results from 2 studies—a registry of 28 sites, and a physician-reported clinical utility trial—reflect real-world experience with CancerTYPE ID. Interim results from the registry study, involving 134 patients, showed that use of CancerTYPE ID changed treatment decisions in approximately 50% of cases.13
The clinical utility study was a retrospective survey-based study of 103 medical oncologists who ordered CancerTYPE ID.18 The primary objectives were to examine whether the results aided in the determination of a clinical diagnosis of the primary site of origin for their patient’s tumor, and the therapeutic decision-making. The assay was used to address a broad range of issues related to diagnostic uncertainty, including independent confirmation of diagnosis prior to initiating treatment, reassessment of diagnosis following unsuccessful treatment, resolution of narrow differential diagnoses, and identification of primary tumor sites in patients diagnosed with CUP.
As Benjamin Kim, MD, MPhil, and colleagues noted, the data reported in this study highlight the clinical impact of obtaining a definitive diagnosis because “the 6 most common final clinical diagnoses (lung, pancreatobiliary, ovary, kidney, breast, and melanoma) require different treatment strategies, and several have approved, targeted therapies available.”
Respondents indicated that the assay provided clinical utility for therapeutic decision-making, with 81% of physicians answering that they “strongly agree” or “agree” that the assay was helpful in the treatment decision-making process.
CancerTYPE ID predicted 26 different common and rare tumor types, and identified a primary tumor type not previously considered in one fourth of the patients. In 41% of the cases, the tumor type identified by CancerTYPE ID was not considered prior to ordering the assay, but the final diagnosis was modified to reflect the test result in two thirds of these cases. Few additional diagnostic tests were ordered after the molecular test, suggesting that most oncologist respondents viewed the results of the assay as being definitive.
Discussion
Early results from basket trials have demonstrated that detecting genetic mutations alone is not associated with treatment benefits from targeted therapy. When applying these data to patients with metastatic cancer of an unknown or unclear origin, it becomes apparent that every effort must be made to achieve a definitive diagnosis before considering targeted therapies based on the identification of mutations alone.
Molecular classifiers are now included in diagnostic algorithms.19 “In practice, the niche for profiling assays are the cases with a poor immunohistochemistry profile and for which 2 completely different therapies are indicated (based on pathology differential), with an impact on quality of life and possibly survival,” according to Varadhachary and Raber.20 “[We] can envision using an integrated algorithm that helps identify a patient’s CUP profile using directed immunohistochemistry and molecular profiling in selected cases.”
CancerTYPE ID helps to achieve a definitive tumor classification for 50 tumor types and subtypes, with an accuracy of 87%. The assay requires minimal tissue—as few as 300 cells—overcoming one of the challenges of standard diagnostic testing (limited tissue availability). The turnaround time is normally within 5 business days and is reimbursed 100% by Medicare. CancerTYPE ID has the unique ability to go beyond the limitations of IHC and accurately differentiate between tumor types in diagnostically challenging cases.
References
- Ciriello G, Miller ML, Aksoy BA, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127-1133.
- Fong TH, Govindan R, Morgensztern D. Cancer of unknown primary. J Clin Oncol. 2008;26(Suppl):22159.
- Thomas SP, Braiteh FS, Cherkis KA, et al. Molecular profiling with the 92-gene assay and decision-impact on cancer treatment: interim results from a prospective, multidisciplinary study. Abstract presented at: 2015 Gastrointestinal Cancers Symposium; January 15-17, 2015; San Francisco, CA.
- Kantar Health, A WPP Company. Epidemiology. www.kantarhealth.com/services/epidemiology. Accessed December 17, 2015.
- Shahda S. Metastatic patients with unclear diagnosis. CME presented at: ASCO Puerto Rico; October 2014.
- Le Tourneau C, Delord JP, Gonçalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16:1324-1334.
- Lopez-Chavez A, Thomas A, Rajan A, et al. Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology phase II basket trial. J Clin Oncol. 2015;33:1000-1007.
- Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;373:726-736.
- Lewis R. Mutation and location important in cancer treatment. Lancet Oncol. 2015;16:e482.
- Kopetz S, Desai J, Chan E, et al. Phase II pilot study of vemurafenib in patients with metastatic BRAF-mutated colorectal cancer. J Clin Oncol. 2015;33:4032-4038.
- Weiss LM, Chu P, Schroeder BE, et al. Blinded comparator study of immunohistochemical analysis versus a 92-gene cancer classifier in the diagnosis of the primary site in metastatic tumors. J Mol Diagn. 2013;15:263-269.
- Anderson GG, Weiss LM. Determining tissue of origin for metastatic cancers: meta-analysis and literature review of immunohistochemistry performance. Appl Immunohistochem Mol Morphol. 2010;18:3-8.
- Laouri M, Schroeder B, Chen E, et al. Diagnostic utility of molecular profiling for cancers of uncertain primary. Abstract presented at: 2011 ASCO Annual Meeting; June 3-7, 2011; Chicago, IL.
- Allen TC. Second opinions: pathologists’ preventive medicine. Arch Pathol Lab Med. 2013;137:310-311.
- Kerr SE, Schnabel CA, Sullivan PS, et al. Multisite validation study to determine performance characteristics of a 92-gene molecular cancer classifier. Clin Cancer Res. 2012;18:3952-3960.
- Hainsworth JD, Rubin MS, Spigel DR, et al. Molecular gene expression profiling to predict the tissue of origin and direct site-specific therapy in patients with carcinoma of unknown primary site: a prospective trial of the Sarah Cannon Research Institute. J Clin Oncol. 2013;31:217-223.
- Hainsworth JD, Schnabel CA, Erlander MG, et al. A retrospective study of treatment outcomes in patients with carcinoma of unknown primary site and a colorectal cancer molecular profile. Clin Colorectal Cancer. 2012;11:112-118.
- Kim B, Schroeder BE, Schnabel CA. Physician-reported clinical utility of the 92-gene molecular classifier in tumors with uncertain diagnosis following standard clinicopathologic evaluation. Pers Med Oncol. 2013;2:68-76.
- Varadhachary GR, Raber MN. Cancer of unknown primary site. N Engl J Med. 2014;371:757-765.
- Varadhachary GR. Carcinoma of unknown primary: focused evaluation. J Natl Compr Canc Netw. 2011;9:1406-1412.

