The description of molecular drivers in diverse cancers, the availability of drugs targeted to these drivers, and the advent of affordable, massively parallel sequencing technology (next-generation sequencing [NGS]) has spurred the oncologic community to develop new paradigms for therapeutic development based on molecular features rather than conventional pathology and a cancer’s site of origin. For some molecular abnormalities, the specific organ in which the tumor developed is an important factor in whether a targeted drug will produce a response or benefit for the patient, while for others, benefit may be seen in different tumors. For example, it is well known that colorectal cancers with BRAF V600E mutations are relatively resistant to BRAF inhibitors, even though these drugs produce responses in most patients with melanoma with BRAF V600E mutations.
1
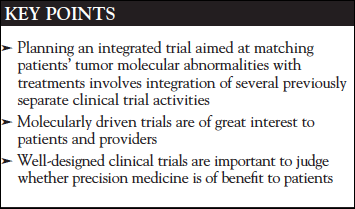
However, there are other examples where a drug may produce responses in various tumors, if the tumors have the appropriate molecular abnormality. Imatinib has been approved by the FDA for chronic myelogenous leukemia in adults and children, where most patients have a BCR-ABL fusion, as well as in acute lymphoblastic leukemia with the same fusion. Imatinib is approved for gastrointestinal stromal tumors, most of which have either KIT or PDGFR mutations.2 The US Orphan Drug Act3 also allowed approval of imatinib based on a basket trial or other data in tumors that have abnormalities of PDGFR or BCR-ABL fusions: myelodysplastic/myeloproliferative diseases, aggressive systemic mastocytosis, chronic eosinophilic leukemia or hypereosinophilic syndrome, and dermatofibroma protuberans.4-6 Likewise, trastuzumab has been found to be effective in combination with appropriate chemotherapy in both breast cancer and gastric/gastroesophageal cancer that overexpress or have amplifications of ERBB2.7,8
Many oncologists consider analysis of somatic genetic alterations to be a standard component of care for advanced cancer patients. However, only a limited number of molecularly targeted therapies are FDA approved in a small percentage of biomarker-defined subpopulations. A far larger percentage of patients will have findings for which there are treatment strategies hypothesized to offer benefit, but for which evidence from clinical trials is still lacking. At present, more patients could be navigated to investigational therapies on clinical trials than to FDA-approved treatments in clinical practice.
Recently, new types of clinical trials have been performed to assess the utility of assigning patients to particular treatments based on the tumor’s molecular profile.9 Umbrella trials usually focus on a single type of cancer, assigning patients who have that type of cancer to experimental treatments based on the molecular profile of their tumor. The National Cancer Institute (NCI)-supported, public-private partnership trial Lung-MAP (www.lung-map.org/about-lung-map) is an umbrella trial. Basket trials, on the other hand, focus on a particular drug or treatment, which is then assessed in patients with various types of malignancy (Figure 1).
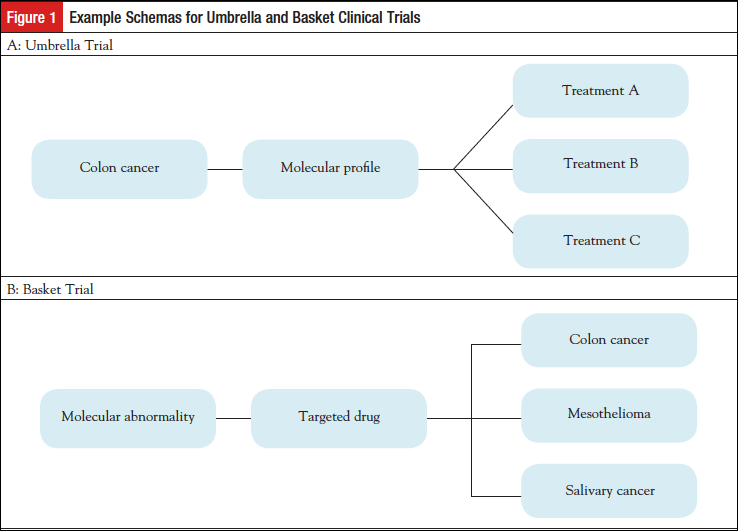
The NCI-MATCH (Molecular Analysis for Therapy Choice; NCT02465060) is a signal-seeking trial that was developed to address the issue of whether a drug, or drug combination, targeted to a specific molecular abnormality could produce responses or prolonged stable disease across the spectrum of cancer types in which the molecular abnormality is found. The trial was designed as a master protocol for molecular screening, with various associated phase 2 subprotocols or “arms” that could be added or closed without affecting the major trial. NCI-MATCH has been referred to as an umbrella-basket, or hybrid trial (Figure 2).
Methods
Planning
To implement the NCI-MATCH trial, NCI partnered with the ECOG-ACRIN Cancer Research Group as the lead National Clinical Trials Network (NCTN) group; all NCTN groups as well as the NCI Community Oncology Research Program (NCORP) could then access and participate in the trial. The NCTN consists of the ECOG-ACRIN Cancer Research Group, Alliance for Clinical Trials in Oncology, SWOG, NRG Oncology, and the Children’s Oncology Group. An important attribute of this NCI-supported system is a cadre of experienced clinical investigators comprising up to 2400 clinical sites, as well as a central ability to ship experimental drugs to the clinic that is treating the patient.
A steering committee was formed consisting of ECOG-ACRIN members, other NCTN members, community oncologists, patient advocates, statisticians, as well as NCI personnel from the Cancer Therapy Evaluation Program (CTEP), the Biometrics Research Program, the Cancer Imaging Program and the Cancer Diagnosis Program within the Division of Cancer Treatment and Diagnosis, and personnel from the Division of Cancer Prevention, which oversees NCORP. Several committees were formed to address critical issues and provide needed content expertise for this first ever type of clinical trial (Table 1). External input was also sought. Project management was implemented, and weekly to monthly meetings were scheduled for each committee. Major tasks included the statistical design for the arms and the overall trial, choosing and contracting for drugs, implementation of analytically and clinically validated integral assays for screening, and developing a rules engine for assigning patients to a particular treatment. We assessed the prevalence of various cancer-related molecular abnormalities from public data sources such as The Cancer Genome Atlas,10 the Catalogue of Somatic Mutations in Cancer (COSMIC),11 cBioPortal for Cancer Genomics,12 My Cancer Genome,13 and various other databases. None of these databases had collected their data in the exact population proposed for NCI-MATCH; eg, patients with solid tumors or lymphomas whose tumor had progressed despite standard therapy, had good performance status, and adequate organ function. However, these databases were crucial for estimating the initial frequency and tumor types in which the driver mutations may be found.
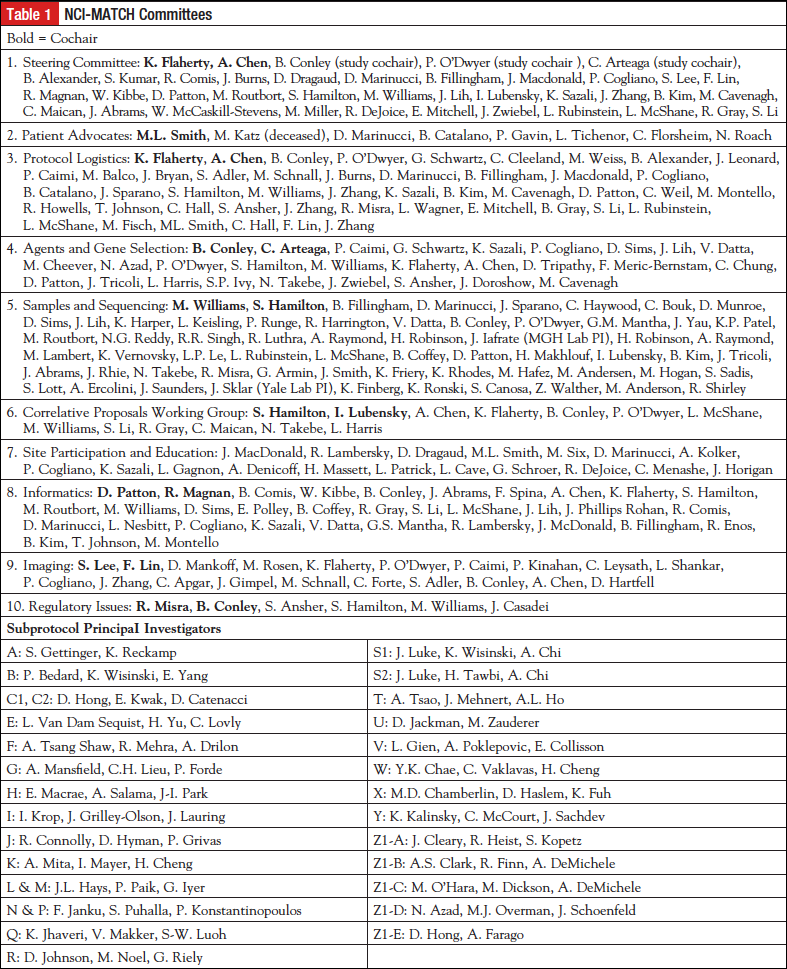
Design
Eligibility for NCI-MATCH was envisioned as patients with solid tumors or lymphoma who had progressed despite standard therapy, and who had good performance status, good organ function, and a life expectancy of at least 3 months. Patients undergo biopsy during the screening period for tumor profiling, because it is unknown if archived tumor contained all the abnormal variants of the current malignancy. The primary end point is overall response rate, as measured by RECIST version 1.114 for solid tumors, the Cheson criteria for lymphoma patients,15 and the RANO criteria for glioblastoma patients.16 For the statistical plan, we considered that for drugs that have recently been approved by the FDA for a given target in a specific malignancy, response rates of 50% to 60% are observed. Because NCI-MATCH would be agnostic to tumor organ site of origin, we postulated that a response rate of 25% would be promising, whereas a response rate of 5% or less would not. This required enrollment of 31 assessable patients per arm. Secondary objectives included assessment of the proportion of patients with time to progression of 6 months or greater, overall survival for each arm, and toxicity. Another goal was to ensure that at least 25% of patients accrued would have less common tumors (eg, other than lung, breast, prostate, or colorectal cancer). This was because the less common cancers may not have enough patients to mount dedicated trials in those diseases, and signals could be generated within NCI-MATCH that might portend patient benefit. The study is monitored for accrual of rarer tumors, in case limits are needed for accrual of the most common cancers. The initial screening goal was 3000 patients, and we anticipated that we might be able to enroll 1000 patients onto a subprotocol, with a goal of about 25 subprotocols covering the most common molecular abnormalities seen in cancer. An interim analysis was scheduled at registration of about 500 patients to assess accrual rate, match rate, types of tumors entered, and to assess for other issues that might impede the success of the trial. We anticipated a period of time of slower accrual, common for NCTN clinical trials, and then an accrual rate of approximately 50 patients/month based on the accrual rates observed in other NCTN clinical trials.
Screening
The concordance of different assays, NGS platforms, and assay procedures is not known. We therefore sought to standardize the procurement of the biopsies, handling, shipping, tumor cell enrichment, and nucleic acid isolation procedures, as well as the NGS assay and other assays used for eligibility for NCI-MATCH. Detailed biopsy procedures were stated in the protocol. Patients could have a low-risk (anticipated serious complication rate <2%) procedure (usually under radiologic guidance) to obtain 4 cores and an optional needle biopsy. Alternatively, tissue from a clinically necessary biopsy could be submitted. Standard biopsy kits were shipped to the enrolling site upon registration of the patient. All biopsies were shipped to MD Anderson, the central receiving laboratory for ECOG-ACRIN, which performed the confirmation of diagnosis and estimation of tumor percentage, macrodissection, and isolation of nucleic acids for shipment to 1 of 4 Clinical Laboratory Improvement Act (CLIA)-certified sequencing laboratories. The goal is to submit specimens that are composed of at least 70% tumor to the sequencing assays. The sequencing laboratories included the Molecular Characterization Laboratory at the Frederick National Laboratory for Cancer Research (P. Mickey Williams, PhD), and 3 competitively chosen CLIA-certified sequencing laboratories: the MD Anderson Molecular Diagnostics Laboratory (Stanley R. Hamilton, MD), the Massachusetts General Hospital Center for Integrated Diagnostics (A. John Iafrate, MD, PhD), and the Yale Clinical Molecular Pathology Lab (Jeffrey Sklar, MD, PhD). In addition, the MD Anderson laboratory performs the nonsequencing assays needed to profile the tumor, currently immunohistochemical (IHC) assays for protein expression of PTEN, MLH1, and MSH2. IHC for retinoblastoma protein expression is performed if a sequencing result reveals high copy number of CCND1, CDK4, or CDK6.
We implemented a validated targeted NGS assay. To do this, it was necessary to choose a platform that could accept very small tumor specimens, such as would be available from sites across the NCTN (academic and large and small community sites) that would be performing the biopsies. The proposed turnaround time (2 weeks) and the uncertain accrual rate demanded a platform that could run with 1 or a few specimens per day. After reviewing results from a Request for Information published in the Federal Record, the NCI chose the Thermo Fisher Ion Torrent Personal Genome Machine for this purpose. The 4 contracted CLIA-certified laboratories validated the same locked assays with excellent concordance (Li J, et al; personal communication, article submitted for publication). Having 4 laboratories would ensure no lag time if 1 of the laboratories had “down-time” for technical or other reasons. The NGS assay detects single nucleotide variants, copy number variants, insertions, deletions, and selected translocations in 143 genes. The intended turnaround time was within 14 days. The NCI-MATCH team met twice with the FDA Center for Devices and Radiological Health in presubmission conferences. Because the study was not considered to be a significant risk, the device is under an abbreviated Investigational Device Exemption, requiring patient consent for and sponsor (NCI) periodic monitoring of the performance and risk of the investigational assay. Laboratory results for the IHC studies and the sequencing are returned to the clinician to be shared with the patient. Recently it has been shown that germline (inheritable) cancer risk mutations may be uncovered by tumor sequencing, even with a targeted NGS assay.17 NCI-MATCH does not sequence germline tissues, but if mutations associated with germline cancer risk are present in the tumor, the laboratory report will alert the clinician to consider the patient’s disease and family history and refer the patient for genetic counseling and assessment for germline mutation as appropriate.
A different committee, the Agents and Genes working group, met with several pharmaceutical companies to develop appropriate arms for the trial, with the intention of including treatments for patients with molecular abnormalities present at a frequency of 2% or greater. The NCI-MATCH team worked closely with CTEP to expeditiously complete contract agreements with the pharmaceutical companies. In addition, “Levels of Evidence” for both the drugs to be included as well as the variants that might serve as eligibility criteria were developed (Table 2). The lowest level of evidence for each treatment arm was that at least 1 patient with the relevant molecular abnormality had been documented to have responded to a drug targeted to that abnormality. The NCI-MATCH team made a concerted search for single agents or combinations of drugs that had promising activity against any known driver of a malignant disease. Any single-agent drug or combination for NCI-MATCH required a recommended phase 2 dose and evidence of clinical activity in at least 1 patient with the relevant actionable mutation. This committee continues to meet on a monthly basis to consider needs for the NCI-MATCH trial, to meet with pharmaceutical companies interested in NCI-MATCH, and to review proposals proffered by clinical investigators interested in including an arm in NCI-MATCH. The drug(s) that met the requirements moved forward to proposal to CTEP for review as a subprotocol of NCI-MATCH if the relevant companies were willing to supply drug. Patients with tumors for which the drug may already have been FDA approved were not eligible for treatment, and patients with tumors for which the treatment had been shown not to have efficacy were also excluded. Pharmaceutical companies were not asked to support screening. The steering committee decided to involve 2 principal clinical investigators and potentially a third translational principal investigator for each subprotocol. The subprotocol investigators were recommended by NCTN members and were tasked with maintaining a deep understanding of which variants had the requisite level of evidence, and with communicating any changes needed to the protocol due to new information about drugs or targets. These investigators were young or mid-career investigators, mentored by experienced NCTN investigators. This procedure allowed each subprotocol the deep expertise needed to keep abreast of advancing evidence, while allowing the subprotocol principal investigators to become familiar with NCTN procedures. ECOG-ACRIN coordinated communications between subprotocol principal investigators, NCI Central Institutional Review Board (CIRB), and others.
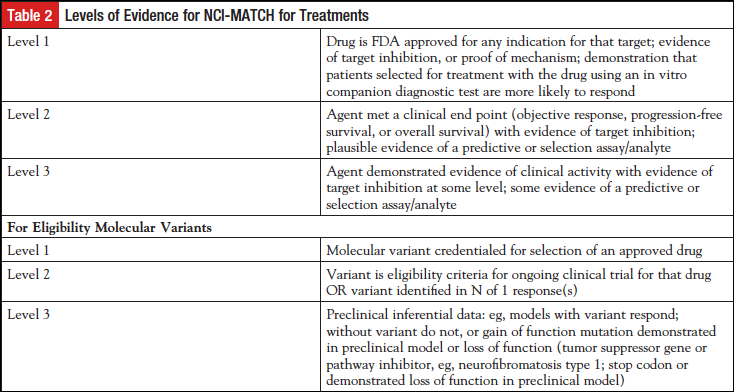
Pharmaceutical companies participate in NCI-MATCH by providing drugs as well as expertise, particularly in assigning actionability to a particular mutant variant. Such expertise often included knowledge of the prevalence of the mutant variant in the population with malignancy. Results (clinical and laboratory) are shared with the pharmaceutical companies under agreements with NCI. Agreements comply with the “NCI IP [intellectual property] option,” whereby the pharmaceutical company supplying the drugs cannot be blocked from using new IP that may be generated in NCI-MATCH.
Informatics
Instead of a tumor board, which may lead to inconsistencies of treatment assignment in practice, an investigational informatics rules engine (MatchBox) was developed that used the molecular variants, plus any other laboratory eligibility assays, the patient’s disease, any prior relevant therapy, and the molecular or previous treatment inclusion and exclusion stipulations in each subprotocol to assign a patient to an NCI-MATCH treatment arm. The results of this assignment were reviewed daily by the MATCH leadership (at least 1 oncologist, 1 laboratorian, and 1 informatics specialist) for accuracy. The rules engine allowed a patient to enter subsequent NCI-MATCH arms as follows: If the patient were ineligible for the initially assigned treatment, or the patient’s disease progressed within 6 months, without a response, and the patient’s tumor had a second actionable mutation, the patient would be offered participation in a second arm of NCI-MATCH. If the patient’s tumor responded, or progressed after receiving at least 6 months of treatment on the original arm, then the patient would be required to have a second biopsy if the patient wished to be considered for a second NCI-MATCH treatment.
Other committees addressed patient and investigator education, public relations, protocol logistics, and imaging. The patient advocate committee assessed the willingness of patients to undergo biopsy to be assigned a treatment. Patients did find a biopsy to be acceptable, given that the risk of serious complications was expected to be less than 2%. The Patient Advocate Committee also provided valuable input in many decisions that guide NCI-MATCH. NCI-MATCH was a new type of trial for the NCTN, and thus, several education sessions were held with the investigators and clinical research staffs of the research sites to ensure the procedures in the protocol were clear and understood. We implemented central contacts for questions from the enrolling sites that could be transmitted directly to trial leadership as needed, as well as to the laboratories.
The trial was reviewed by the NCI CIRB. All participating sites were required to accept the CIRB as the IRB of record. This requirement was necessary because the rules engine only allowed the current arms to be considered for assignment of treatment, and a site could not have a different number of arms than the currently active version of NCI-MATCH.
Activation and ImplementationBecause it was not feasible to activate all arms simultaneously, the study was reviewed by the CIRB with 4 arms, and “preactivation” was allowed by the sites to prepare them for trial opening. The trial opened once 10 arms were approved by CIRB and CTEP, with the intention of adding arms as the study progressed.
Results/Lessons LearnedA signal-seeking trial such as NCI-MATCH is of interest and acceptable to patients and clinicians. The NCI-MATCH accrued nearly 800 patients for molecular screening in the first 3 months. This implies a willingness to participate in such trials and potentially enthusiasm to assess targeted agents in any malignancy that possessed the relevant mutation. However, we have yet to learn more about the correct timing in a patient’s disease course to do this type of tumor profiling. We do hope that many patients will agree to provide archived specimens for comparison to the fresh specimen provided for NCI-MATCH, so that we can better understand whether an archived specimen will suffice, or whether it is necessary to obtain a more proximate tumor specimen.
The trial opened in August 2015 with 10 arms. At that point, the expected match rate was about 10%, based on estimates from accessible databases (see above). The interim analysis will be described in a separate publication. The intention was to add more arms during the “ramp up” period. However, at the interim analysis, and largely because of the tremendous enrollment rate, the trial did pause for new enrollment and was reopened to new enrollment May 31, 2016, with 24 arms (Table 3). During this pause, any assigned patients continued on treatment, and, as new subprotocols were added, any patients who were sequenced prior to the pause and had mutations matching the new arms were offered those treatments.
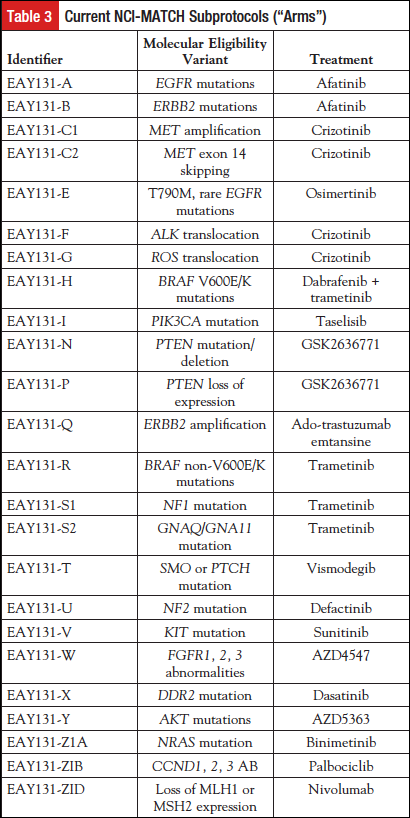
Tumor profiling as a national activity is feasible. More than 85% of the biopsies submitted were profiled successfully, consistent with the experience of several major academic medical centers that routinely perform similar NGS analyses. However, this did require the central laboratory to perform enrichment macrodissection, sometimes extensively. As a result, turnaround times suffered, and the enthusiastic uptake of the trial called for the deployment of significant additional laboratory resources.
Mounting such a trial requires more resources than expected. Laboratory resources needed to be increased, as noted above. NCI-MATCH required 10 subcommittees; in all, over 150 professionals committed significant time over a period of 3 years to make the trial a success. However, this work was coordinated, sustained, and cooperative. It is essential to learn if the practice of tumor profiling in patients who have progressed after standard treatment is capable of providing benefit to patients. Trials such as NCI-MATCH present a tremendous opportunity to learn about mutations present in tumors seen by oncologists in the community, as well as in academic centers. As such, this trial represents a chance to both catalog the mutations and to test their responsiveness to targeted treatment. NCI-MATCH is intended to be flexible so that if a signal (response) is seen in a rare tumor, that arm could be amended to include a statistically determined number of patients with the same tumor and same mutation. If new treatments are developed or new actionable mutations are discovered, then MATCH could add new arms to address these.
NCI-MATCH may provide a platform for future drug development. It may be possible to add pharmacodynamics end points within such a trial, as well as to do limited dose finding for previously unexplored combinations. As has been published, there are a number of driver mutations that exist at low frequency in the population, and there may be treatments that could be greatly beneficial for the 5% or less of patients who have them. In addition, this type of clinical trial platform could explore treatments in particular tumor types where targeted therapies have been somewhat less successful, or where the targeted mutation occurs in a very low proportion of the patients. A large screening effort is necessary to find such mutations. One way to explore a possible signal in a particular tumor tissue type is to have expanded cohorts in each arm that address a relevant mutation for that particular tumor (eg, colorectal cancer). A relatively small cohort of such patients (about 15) could be accrued to assess whether a sufficient response signal is present in an individual tumor type with a particular mutation. An insufficient response may prevent mounting trials that will eventually fail (likely involving a fairly massive screening effort), whereas a positive signal may move the treatment closer to FDA approval.
It is still a minority of patients with advanced refractory cancer who undergo targeted sequencing and other tumor profiling who are able to be matched with an actionable mutation. Trials have been reported that have been unsuccessful in proving that screening for molecular mutations can be beneficial to patients,18,19 whereas others have reported increased responses using such an approach.20 While we are aware of these results, the research community must continue to do studies to understand what is necessary for success, if it is possible. With the addition of arms to NCI-MATCH, we expect to be able to match 20% to 30% of such patients, but not all matched patients will actually be able to be treated due to various factors, such as safety eligibility criteria relevant to organ function, cardiac function, declining performance status, or current or prior drug treatment. Nevertheless, it is important to accomplish trials such as NCI-MATCH to understand if there is a good response rate for patients who do get treated under our levels of evidence. In addition, we expect, even in a cohort of patients with similar mutations, treated similarly, that not all will have a response. We can use the tumor profiling results, and possibly whole exome sequencing and RNA sequencing, to better understand the molecular reasons for response or nonresponse to matched targeted treatments.
DisclaimerDr Conley is an employee of the National Cancer Institute. However, the opinions expressed by Dr Conley are her own, and not necessarily those of the National Cancer Institute, National Institutes of Health, or the US Department of Health and Human Services.
Potential Conflicts of InterestBarbara A. Conley: None
Keith T. Flaherty: Consultant to Genentech, Novartis; funded research: Novartis.
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100-103.
- Cioffi A, Maki RG. GI stromal tumors: 15 years of lessons from a rare cancer. J Clin Oncol. 2015;33:1849-1854.
- Orphan Drug Regulations, Final Rule. Food and Drug Administration, Department of Health and Human Services. Federal Register. 2013;78(113):35117-35135.
- Tefferi A, Pardanani A. Imatinib therapy in clonal eosinophilic disorders, including systemic mastocytosis. Int J Hematol. 2004;79:441-447.
- Duffaud F, Le Cesne A. Imatinib in the treatment of solid tumours. Target Oncol. 2009;4:45-56.
- Rutkowski P, Przybył J, Switaj T. Genetics of rare mesenchymal tumors: implications for targeted treatment in DFSP, ASPS, CCS, GCTB and PEComa. Int J Biochem Cell Biol. 2014;53:466-474.
- Puglisi F, Fontanella C, Amoroso V, et al. Current challenges in HER2-positive breast cancer. Crit Rev Oncol Hematol. 2016;98:211-221.
- Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687-697.
- Siu LL, Conley BA, Boerner S, et al. Next-generation sequencing to guide clinical trials. Clin Cancer Res. 2015;21:4536-4544.
- The Cancer Genome Atlas Network; Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113-1120.
- Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945-D950.
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-404.
- My Cancer Genome. www.mycancergenome.org. Accessed August 18, 2016.
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228-247.
- Cheson BD, Fisher RT, Barrington SF, et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol. 2014;32:3059-3068.
- Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963-1972.
- Meric-Bernstam F, Brusco L, Daniels M, et al. Incidental germline variants in 1000 advanced cancers on a prospective somatic genomic profiling protocol. Ann Oncol. 2016;27:795-800.
- Le Tourneau C, Delord JP, Gonçalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled, phase 2 trial. Lancet Oncol. 2015;16:1324-1334.
- Lopez-Chavez A, Thomas A, Rajan A, et al. Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology phase II basket trial. J Clin Oncol. 2015;33:1000-1007.
- Tsimberidou AM, Wen S, Hong DS, et al. Personalized medicine for patients with advanced cancer in the phase I program at MD Anderson: validation and landmark analysis. Clin Cancer Res. 2014;20:4827-4836.
Dr Conley is Associate Director of the Cancer Diagnosis Program in the Division of Cancer Treatment and Diagnosis, National Cancer Institute (NCI). She is also a member of the division’s experimental therapeutics clinic. Within the NCI, she leads the NCI Molecular Analysis for Therapy Choice (NCI-MATCH) trial and the Exceptional Responders Initiative.
Dr Flaherty is Director, since 2012, of the Henri and Belinda Termeer Center for Targeted Therapy and, since 2014, Director of Clinical Research at the Massachusetts General Hospital and Professor of Medicine at Harvard Medical School.

