The Third Annual PMO Live Conference, a Global Biomarkers Consortium Initiative, took place in San Francisco, California, on October 31 – November 1, 2014. PMO Live is the only global meeting dedicated to advancing the understanding of value and clinical impact of biomarker research in oncology. Following are highlights of presentations from the meeting.
New Diagnostic Approach Identifies
Actionable Genetic Alterations in Tumors
Missed by Traditional Tests
The utility of traditional tests to identify actionable genetic alterations in tumors is suboptimal, whereas comprehensive genetic profiling can identify clinically meaningful alterations in 85% of cancer patients, said Juliann Chmielecki, PhD.
As more clinically relevant cancer genes are identified, comprehensive diagnostic approaches are needed to match patients to therapies.
Molecularly targeted therapy is evolving, but the number of clinically relevant cancer genes across solid tumors is high. In 2012, Stephens et al found at least 1 clinically relevant genomic alteration in 59% of colorectal cancer (CRC) and non–small cell lung cancer (NSCLC) tissue specimens, and revealed 2 gene fusions: C2orf44-ALK in a CRC and KIF5B-RET in a lung adenocarcinoma. Further screening of 561 lung adenocarcinomas found RET fusions in 2%.
The pace of targeted therapy discovery is rapid, such that by 2014, RET fusion–positive NSCLC was found to respond to a RET inhibitor (cabozantinib).
Multiple different diagnostic tests for the myriad of relevant genetic alterations in cancers may exhaust precious biopsy material, said Chmielecki, senior scientist of cancer genomics at Foundation Medicine. The low tumor purity in many clinical tumor specimens is a diagnostic challenge that requires diagnostic tests with high accuracy. (Purity is the relative proportion of extracted DNA originating from tumor cells.)
In a study of clinically relevant somatic mutations in NSCLC specimens, the fraction of mutations with an allele frequency under the limit of detection by capillary Sanger sequencing was 55%. “Next-generation sequencing–based tests miss most of the actionable alterations within a given patient,” she said.
Given the challenges of small biopsies, multiple classes of genetic alterations, and tumor samples that may have small tumor content, “we developed a test that has all 4 classes of alterations with very high sensitivity and positive predictive value,” she said. The cancer genomic profiling test is based on massively parallel DNA sequencing to characterize base substitutions, short insertions and deletions, copy number alterations, and selected fusions across 314 cancer-related genes from fixed formalin paraffin-embedded specimens.
Experience in more than 13,000 patients who had genomic profiling of their tumors shows 97% with an alteration reported and 85% of samples with at least 1 clinically relevant alteration. Clinical relevance was defined as an alteration for which an FDA-approved targeted therapy in the tumor type or in another tumor type existed, or an open clinical trial of therapy relating to the alteration found in the gene.
“When we look at the 314 cancer-related genes, we’re not finding a huge number of alterations per patient,” she said. “A higher number is usually cancers that are associated with carcinogens, such as lung cancer and melanoma.” The mean number of alterations per sample was 4.5 (range, 0-57), and the mean number of clinically relevant alterations per sample was 2.0 (range, 0-28).
Solid tumor types represented in the more than 13,000 clinical specimens included lung (19%), breast (14%), colon (9%), brain (5%), ovary (5%), pancreas (4%), soft tissue (4%), and other (32%; mainly rare tumors).
The investigators found a huge number of low-frequency alterations that could be linked to targeted therapies “that may be clinically relevant, even though they’re not occurring in hot spots of high-frequency genes,” she said.
A novel KIF5B-RET fusion was identified and later discovered to be in 1% to 2% of NSCLCs; KIF5B-RET transformed cells are sensitive to RET inhibitors, suggesting that RET kinase inhibitors should be tested in prospective trials in patients with NSCLC with RET fusions. Multiple patients with KIF5B-RET fusion are known to have responded to a RET inhibitor, she indicated.
HER2/ERBB2 alterations were observed across 27 tumor types. “This phenomenon may extend well beyond the gastric and breast cancer fields and into areas that haven’t been looked at,” said Chmielecki. “There are clinical case reports showing sensitivity of HER2 amplification outside of breast and gastric cancer as well as some of these newer mutations being discovered.” Nearly half of all HER2/ERBB2 alterations are missed by current tests, she noted.
Epidermal growth factor receptor (EGFR) alterations were identified in 151 cases (6.8% of total cases) across 13 different tumor types, including breast cancer. An EGFR mutation is not usually tested for in a person with breast cancer. By site of organ, EGFR alterations occurred in glial, lung, brain, head and neck, bladder, kidney, uteral, breast, bone, colorectal, esophageal, skin, and stomach tumors.
Fusions were identified in 13 kinases across multiple tumor types. Of the 248 kinase fusions identified, many with novel 5??partners were identified (2.2% of total cases).
Payers Debate Economics and Molecular Biomarkers in Personalized Medicine
The rising costs of biomolecular testing and targeted drugs have prompted many to ask whether the United States can afford personalized medicine in oncology. At the conference, medical directors from 2 health plans tackled this question from the payer perspective.
Ken Schaecher, MD, medical director at Select-Health, answered “it depends” to the key question. He noted that healthcare costs in the US now stand at 17.2% of the gross domestic product. The US can afford personalized medicine “until we decide not to,” he said, pointing out that some targeted medicines cost upward of $100,000 per year because the US has no threshold to pay for these medicines.
Most payers have defined sets of rules, he said, but the problem in the US is the large number of payers, each with its own set of rules. Sometimes, even within a plan, one set of rules may be applied, but different benefits structures mean that one patient can get a therapy that another cannot.
Gary Johnson, MD, MS, MBA, regional medical director at Humana, said that paying for treatments and tests is always possible, but the real question is, “should we pay?” As stewards of the premium dollar, insurers can cover anything they want to, but they’ll have to raise premiums to do so.
The targeted approach to care allowed by personalized medicine creates the opportunity for greater cost-effectiveness, as in KRAS testing to select patients for targeted therapy in CRC. Being able to select the right treatment for the right patient improves health outcomes, said Johnson.
“It [personalized medicine] shows us where treatments will be effective, and just as important, where treatments will not be effective, so we can avoid toxicities and morbidities from treatments that in essence cause harm,” he said. “All of this is the goal of managed care and of clinical care.”
By reducing unnecessary risks of treatments not likely to work in a given patient, personalized medicine can reduce liability, said Johnson. A well-defined treatment pathway linked with a diagnostic marker provides legal cover for denial of a treatment.
“We can identify disease at an earlier state, and as a general rule, the earlier we treat various cancers the more effective it is clinically, and as important, the more cost-effective it is,” he said. “BRCA testing, which has been around for decades, and Oncotype Dx for breast cancer are additional examples of why the answer to this question, can we afford personalized medicine?, is a resounding yes.”
The higher price tags of targeted therapies reduce the cost-effectiveness of personalized medicine, said Schaecher.
“We’ve found that typically with more targeted therapies, the price tag goes up, and there’s only so much money in the bank,” he reasoned. “A great example is crizotinib. It has a $96,000 price because it only works in 4% of the non–small cell lung cancer population that has the mutation. That price tag is much above most of the other therapies used in NSCLC.”
Biomarker costs are increasing as well, he said, with some tests exceeding $3000.
The misapplication of information in directing therapy is another potential con to personalized medicine in oncology. Sometimes, tests with low sensitivity or specificity are being used to determine clinical utility. And in some instances, providers select the same treatment they would have otherwise without the biomarker test.
There is also a risk that information from testing may be applied when evidence is not present, said Schaecher. “It’s arguable that there is adequate evidence for whole genome sequencing in oncology, but for most payers, they don’t believe that the evidence is there yet,” he said. “Using biomarkers in a one-off circumstance can help 1 patient, but unfortunately, as payers, we have to make broad payer-based decisions and not individual decisions. When there’s a lack of evidence, we have to be consistent in our decisions. If I make an exception for one patient, that can bind us to make an exception for every patient going forward.”
Pharmacogenetic testing to identify drug metabolism may also add cost without utility. It’s unproven that knowing that a patient may be a rapid metabolizer of an antidepressant medication is going to alter the therapy. That can occur in the oncology arena also, said Schaecher.
A lack of process standardization across laboratories argues against wholesale use of biomarker tests. “As much as we would like all of the tests to be FDA approved, sometimes FDA approval can mean different things,” he said. “Many [tests] are home brewed, done in university labs, and when you don’t have that standardization, it’s harder for us to want to approve having a test covered.”
Finally, a patient may still insist on a therapy anyway with a negative biomarker test.
“Provide payers with evidence that it’s useful and that it would be no more costly or preferably less costly than the current approach to management, and you win the day,” said Schaecher. “It’s that easy.”
CoMMpass Study Will Drive Myeloma Into Precision Medicine Territory
A landmark clinical trial is underway in multiple myeloma, sponsored by the Multiple Myeloma Personalized Medicine initiative (MMPM) of the Multiple
Myeloma Research Foundation (MMRF). The trial aims to elucidate the molecular variations that underpin the development and progression of myeloma and to create an unprecedented data set to facilitate clinical trials and support the personalized care of patients.
The CoMMpass (Relating Clinical Outcomes in Multiple Myeloma to Personal Assessment of Genetic Profile) trial was described by Jeffrey Wolf, MD, director of the myeloma program at the University of California San Francisco Helen Diller Family Comprehensive Cancer Center.
“The CoMMpass longitudinal study is a core element of the MMPM program to identify patient segments based on their molecular profiles,” Wolf said. He noted that MMRF can already be credited with first identifying a number of unusual mutations in this disease.
The CoMMpass Study will enroll at least 1000 newly diagnosed, symptomatic myeloma patients from at least 90 community and academic cancer centers. These patients will be tracked from diagnosis through treatment over 10 years by means of sequential tissue sampling, mutational analysis, and clinical exam. The goal is to determine how their molecular profiles may affect treatment response and disease progression. Clinical and genomic data will be obtained every 6 months and incorporated into the MMRF Researcher Gateway. The gateway will allow researchers outside of the CoMMpass Study to access the data acquired in the trial.
Centralized molecular tests will include flow cytometry for determining BRAF mutation status, RNA sequencing expression analysis, whole exome DNA sequencing, whole genome chromosome analysis, and cytospin slides for fluorescence in situ hybridization. There will be a biorepository of peripheral blood mononuclear cells, plasma, and tumor tissue.
The study is on track to meet its goals. The current status of projects is shown in the Table.
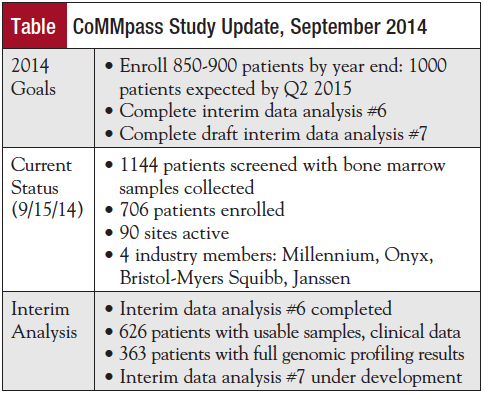
“The CoMMpass Study is already generating high-quality data and achieving critical milestones,” Wolf observed.
By interrogating the molecular profile of tumors and determining whether certain profiles respond best to certain drugs, the CoMMpass Study will help change the paradigm of drug development and treatment from one of “blockbusters” to one of “orphans,” Wolf predicted.
Today, clinical trials aim to test for safety in phase 1, test for activity in phase 2, and compare new treatments to standard treatments in phase 3, he noted. Tomorrow’s will be biomarker-driven Bayesian trials and will essentially be “trials” of individual patients. Results from such trials, he said, will propel the field of multiple myeloma beyond “personalized” medicine and into the “next big leap of precision medicine.”
Progress Is Steady in Search of Molecular Biomarkers for Early Detection of Several Types of Cancer
Of the nearly 5000 molecular biomarkers for early detection, diagnosis, and prognosis of cancer, very few receive regulatory approval. At the conference, a panel informed attendees about the progress made in the development of early detection biomarkers for cancer.
Sudhir Srivastava, PhD, MPH, spoke about developing and validating biomarkers through the Early Detection Research Network (EDRN).
The organizational structure of EDRN is modeled after the drug discovery pipeline and cooperative groups. Funding is provided for individual and collaborative research under coordination and oversight by the National Cancer Institute (NCI). Key elements include biomarker developmental laboratories, reference laboratories for analytic validation, and centers for subsequent clinical validation. “The goal is to accelerate development of useful biomarkers, but also, importantly, to discard poor markers early on, thus avoiding wasted efforts due to false leads and irreproducible early results,” said Srivastava, chief, Division of Cancer Prevention at NCI.
A steering committee of the investigators provides internal governance, and an EDRN consulting team provides independent scientific guidance and review.
Key objectives of EDRN are to establish an investigator-initiated infrastructure that will foster interaction among stakeholders; standardize biomarker validation criteria; and, ultimately bring biomarkers to clinical use. EDRN has 4 major collaborative disease-oriented groups: prostate, lung, colon, and breast and related cancers.
Five FDA-approved and 11 CLIA-approved biomarker tests have been developed from the structure and process.
He provided an example of markers that were tested and compared with fecal occult blood testing and fecal immunochemical test (FIT), and the decision rules, including performance bars, used during the validation process. Of 21 markers tested, 2 markers met the criteria for further evaluation: galectin-3 ligand and methylated vimentin. “Both have far better operating characteristics than CEA [carcinoembryonic antigen], which appears in the middle of the pack,” said Srivastava.
Early Detection of Colorectal Cancer
The rationale for a stool DNA test to detect CRC, and the pivotal study to secure its approval, were discussed by Barry Berger, MD.
The biologic rationale for using stool DNA for a screening test is that colonocytes are continuously shed into the fecal stream, degenerate, and release aberrant DNA biomarkers, he said. Further, colorectal neoplasia-associated biomarkers have been well characterized and are available for analysis.
“Exfoliation of cellular material is scarce in normal colon lining and abundant in advanced adenoma and colorectal cancer,” said Berger, chief medical officer, Exact Sciences Corporation, Madison, WI.
A multitargeted stool DNA test that consists of quantitative molecular assays for KRAS mutations, aberrant NDRG4 and BMP3 methylation, and ß-actin, plus a hemoglobin immunoassay was developed and validated. From quantitative measurements of each marker, a validated, prespecified logistic regression algorithm was derived, with a value of 183 or more indicating a positive test result.
In 9989 asymptomatic individuals at average risk for CRC, the study sought to determine sensitivity and specificity of the multitargeted stool DNA test for CRC and advanced adenoma and to compare the sensitivity and specificity with FIT.
The sensitivity for cancer (92.3% vs 73.8%; P=.0018) and advanced adenoma (42.4% vs 23.8%; P<.0001) was superior to that of FIT, although specificity was lower with DNA testing, said Berger. The stool DNA test was superior in detecting stage I to III CRCs and high-grade dysplasia compared with FIT.
Centers for Medicare & Medicaid Services coverage of the stool DNA test was approved on October 9, 2014.
Airway Biomarkers for Lung Cancer
Bronchial airway gene expression can serve as an early diagnostic biomarker for lung cancer, said Avrum Spira, MD, MSc. Nasal gene expression may serve as a less invasive surrogate for the bronchial airway.
In smokers, epithelial cell gene expression is altered throughout the respiratory tract, but the epithelial cell genomic response to smoking is variable. A validation study of airway epithelial gene expression in 350 smokers undergoing bronchoscopy for suspected lung cancer had a sensitivity of 88% for lesions <3 cm and 86% for early-
stage lung cancer.
Patients with lung cancer have deregulation of 7 different pathways in bronchial epithelial cells, said Spira, from the Division of Computational Biomedicine at Boston University. Gene expression profiling reveals increased activation of PI3K in the airway of smokers with dysplasia, and chemoprevention has been shown to alter airway gene expression in such patients.
The field of injury with smoking extends to the nasal epithelium for both diagnosis and screening, he said. Examination of RNA obtained from nasal mucosal brushing has discovered that nasal gene expression reflects the bronchial airway gene expression response to smoking. Genes associated with cancer in the nasal epithelium are similarly up- and down-regulated in the bronchial epithelium, he said.
MicroRNA regulates part of the gene expression response to smoking. Using small RNA sequencing, a novel microRNA – miR-4423 – was found to be associated with lung cancer and might be useful for early detection. miR-4423 is expressed almost exclusively in the respiratory tract and localizes to airway epithelium, said Spira, and is down-regulated in lung cancer and in the proximal airway of smokers with lung cancer.
Overexpression of miR-4423 has been shown to inhibit tumor cell growth in a subset of cancer cell lines and in a xenograft model.
Early Detection Biomarkers for Breast Cancer
Multiple protein and autoantibody biomarkers have been identified for early detection of breast cancer, said Karen Anderson, MD, PhD.
Antibodies are a measure of the immune response to cancer. An advantage to antibodies in cancer detection is their long half-life and minimal hourly or daily fluctuation. They are stable in a serum sample, said Anderson, associate professor, Biodesign Institute at Arizona State University, Tempe.
Her group probed novel high-density custom protein microarrays expressing nearly 5000 candidate tumor antigens with sera from patients with early-stage breast cancer. From these microarrays, a 28-autoantibody panel for breast cancer detection was identified that had a sensitivity of 80.8% and a specificity of 61.6%, primarily for estrogen receptor–positive cancers.
Triple-negative breast cancer is an especially aggressive form of breast cancer that is often not detected with screening mammography. “The goal for screening is a blood-based test that will lead to further imaging,” she said. Plasma biomarkers are currently undergoing a validation study by NCI/EDRN in an effort to find a composite biomarker panel with 98% specificity and 60% sensitivity for early detection of triple-negative breast cancer.
Cancer Research and Treatment in the Era of Molecular Biomarkers Incorporates New Sequencing Technologies, Clinical Trial Designs
Transformative changes in cancer therapy will require new models for clinical research and practice. Retrofitting current knowledge into traditional paradigms is suboptimal and will slow the progress in discovering effective targeted agents, said Razelle Kurzrock, MD, director, Center of Personalized Therapy and Clinical Trials, University of California, San Diego, Moores Cancer Center.
Common cancers are difficult to treat because each may be >100 different diseases, especially in the metastatic setting. Metastatic tumors are complicated, she said, and it’s unlikely that monotherapy will produce prolonged responses or complete remission.
“While it is often claimed that we need new drugs to treat cancer, a more fundamental problem may be the way we classify cancer,” she said. Although excellent anticancer drugs may exist, they often work poorly because patients with the wrong cancers are being treated with them. In unselected populations, the survival gain with targeted agents is minimal – an average gain of 2 months. These agents work only in those patients with a sensitizing aberration.
More rational patient selection will identify those patients with the best chance to respond, and it is already occurring. The Profile Related Evidence to Determining Individualized Cancer Therapy (PREDICT) Program in Advanced Cancer Patients uses a histology-independent targeted approach in which multiple
molecular aberrations are assessed and used to match patients with targeted agents. Patients with 1 genetic mutation discovered had a 27% complete/partial response rate when a matched therapy was available, compared with only 5% in patients with a cancer that did not have a matching therapy (P<.0001).
“Molecular aberrations do not segregate well by organ of origin,” said Kurzrock.
For example, in PREDICT, a PIK3CA mutation was evident in 10% of advanced cancers originating in various organs, including 29% of endometrial cancers, 24% of breast cancers, and 13% of lung cancers.
Even when initiated in a phase 1 clinical trials program, personalized medicine improves clinical outcomes. At MD Anderson Cancer Center, failure-free survival improved with phase 1 matched therapy but not unmatched compared with prior conventional therapy (Tsimberidou AM, et al. Clin Cancer Res. 2012;18:6373-6383).
Using next-generation sequencing of 75 cancer patients, no 2 patients had the same molecular portfolio, “which speaks to the need to customize therapy for each patient,” she said. “This is going to get even more complicated because so far we’ve only done targeted next-generation sequencing in the genomics, not having touched on transcriptomics, proteomics, or epigenetic changes.”
Three-dimensional in silico modeling is being used to predict response to treatment and is showing that patients with the same genetic abnormality may behave differently depending on where the precise mutation is located. For example, patients with insertion exon 20 mutations are classically resistant to EGFR inhibitors, but 2 patients with NSCLC with insertion exon 20 mutations have responded to cetuximab, which was predicted by in silico modeling “because that mutation brings the dimerization domains closer together.”
Transformation in Chronic Myelogenous Leukemia
The revolution in improved outcomes (>90% response rates, median survival of 20+ years) in chronic myelogenous leukemia (CML) may be a model for solid tumors, Kurzrock said. This revolution has been driven by a known target (Bcr-Abl), development of a targeted agent (imatinib), and a move to treat newly diagnosed patients. When in blast crisis, however, the response rate in CML is <10%, and the median survival is only about 12 months, much like outcomes with solid-tumor metastatic disease.
Strategies to transform outcomes in solid tumors may lie in treating newly diagnosed disease and using combination treatments for advanced disease. Supercomputers are being used to identify convergence pathways in patients with several genetic alterations to reduce the number of drug combinations that may be necessary. Liquid biopsies (sequencing DNA taken from the bloodstream) are also being tried, and “may obviate the problem that each metastasis may be a little different,” she said.
Looking Downstream
The complexity of the molecular pathways and tumor hypermutability and heterogeneity are prompting the search for effective molecular targeting sites downstream from the activated pathways, said Caroline Robert, MD, PhD.
The cap-dependent translation initiation complex appears to be one such promising target, said Robert, head of dermatology, Institut Gustave Roussy, Paris.
When BRAF is inhibited with dabrafenib, the signaling pathway is blocked, resulting in an arrest of cell proliferation and induction of apoptosis. Duration of response to BRAF inhibition is limited, however, due to secondary resistance. Initiating therapy with a BRAF inhibitor and a MEK inhibitor can improve the response compared with a BRAF inhibitor alone, as demonstrated in the phase 3 COMBO-v trial, which demonstrated a 31% reduction in the hazard for survival in patients treated with dabrafenib and trametinib versus vemurafenib monotherapy (Robert C, et al. ESMO 2014. Abstract LBA4). Treatment with the combination was associated with a 44% reduction in the hazard for progression or death.
Most frequently, mechanisms that result in resistance to dabrafenib involve a reactivation of the MAP kinase pathway. A persistence in the formation of the eIF4F complex, a cap-dependent translation initiation complex, is associated with resistance to anti-BRAF therapy. The eIF4E cap-binding protein is part of the eIF4F preinitiation complex. Reducing the level of this translation initiation factor suppresses malignancy.
The formation of the eIF4F complex is associated with multiple mechanisms of resistance to vemurafenib and anti-MEK inhibitors. Inhibiting the eIF4F complex synergizes with inhibiting BRAF to inhibit cell proliferation and tumor growth and may therefore represent a rational therapeutic target in cancers with initial sensitivity to a BRAF inhibitor.
Validating Molecular Biomarker Testing
As sequencing technologies evolve rapidly, ensuring the validity of next-generation sequencing techniques is paramount for clinical decision making, said Mark Sausen, PhD, director of research and development at PGDx, a provider of advanced cancer genome analysis and testing services in Baltimore, MD.
A characteristic acquisition of somatic mutations occurs throughout tumorigenesis. Even within different histologic subtypes of any given tumor type, genetic differences can be observed. Many mutations occur very early in tumor development, necessitating evaluation of the genetic landscape of a patient’s tumor both when initially diagnosed as well as throughout the progression of the disease, said Sausen.
Mutations can come in the form of sequence mutations as well as copy number changes and translocations. Next-generation sequencing allows evaluation of all types of somatic mutations within a single platform. Technical process elements of quality management to ensure analytic validity have been developed by the Next-generation Sequencing: Standardization of Clinical Testing workgroup (Gargis AS, et al. Nat Biotechnol. 2012;30:1033-1036). The validation component to next-generation sequencing–based approaches involves several steps:
- Assay validation – documentation of the assay performance metrics and bioinformatics pipeline
- Quality control – evaluation of metrics over time, ensuring consistency with values provided during validation testing
- Proficiency testing – independent evaluation of performance metrics
- Reference materials – samples used for the evaluation of performance metrics, ideally using disease-associated variants
Performance characteristics include accuracy, precision, analytical sensitivity, analytical specificity, reportable range, and the reference range.
An assay developed by PGDx, called Cancer Select-R, is an assay for detection of sequence mutations, copy number changes, rearrangements, and microsatellite instability in 120 genes. “Information contained within the targeted gene panel reports includes the genetic mutations identified, the clinically actionable alterations, and the performance metrics and limitations of the next-generation sequencing assay employed,” said Sausen.
Examining Circulating Tumor DNA
Liquid biopsy approaches are becoming appealing as a noninvasive way to evaluate a patient’s genotype for sensitizing agents for specific targeted therapies as well as the detection of resistance mutations and the potential for relapse, said Sausen. Circulating tumor DNA was recently found to be detectable across tumor types with rearrangements and sequence mutations (Bettegowda C, et al. Sci Transl Med. 2014;6:224ra24). “In the majority of patients, we were able to detect quantifiable amounts of circulating tumor DNA,” he said. “In some tumor types we could detect it in nearly all patients, and in other tumor types, particularly the brain tumor types, we could only identify circulating tumor DNA in a small subset of patients. Both within and across tumor types, there was a lot of variability in the actual levels of circulating tumor DNA.”
Circulating tumor DNA could be found in early- and late-stage disease in most patients, and, as expected, the fraction of patients with detectable circulating tumor DNA increases as the cancer stage increases.
Tsunami of Clinical Trial Designs Coming
There is a “tsunami” of innovative clinical trial designs testing the effect of matching genomic abnormalities to molecularly targeted therapies, said John J. Wright, MD, PhD.
This tsunami has emerged from the convergence of transformative technology “with the abject failure of our standardized randomized controlled approach using unselected ‘all-comer’ populations,” he said.
The new direction in clinical trial design is converging on platform-based trials, discarding the all-comer trials for a randomized screening-based approach with broad panels of genomic predictors as a definitive new way into thinking how to best integrate the development of targeted drug and biomarker combinations, said Wright, of the Investigational Drug Branch, NCI,
National Institutes of Health, Bethesda, MD.
“These trials may extend the screening technology beyond the pretreatment information by incorporating a repeat biopsy at the time of progression on treatment,” he said.
He described the Adjuvant Lung Cancer Enrichment Marker Identification and Sequencing Trials (ALCHEMIST), a near-term National Clinical Trials Network–based phase 3 master protocol in adjuvant lung cancer. The protocol will screen up to 8000 patients with early- stage, completely resectable or resected, nonsquamous NSCLC.
In the screening component trial, which is the only point of entry for all enrollments, eligible patients will have tumor tissue screened for EGFR mutations and ALK rearrangements in a central CLIA-certified laboratory. Those positive for EGFR mutations or ALK rearrangements will be stratified into 1 of 2 randomized treatment marker–specified clinical trials in which the use of erlotinib and crizotinib will be evaluated. Patients negative for these mutations will be followed for 5 years. All patients will undergo biopsy at disease progression to assess for actionable mutations. Patients will also contribute tissue and blood for genomic research by the NCI,.
“ALCHEMIST is designed to accommodate evolving clinical science and research opportunities,” said Wright. “Evaluation of other targeted therapies could be added to the research effort in the future as new and promising therapies emerge.”
Acquired Resistance to Targeted Therapy in Colorectal Cancer Is Similar to Primary Resistance
KRAS mutations are often drivers of acquired resistance to EGFR inhibitors in the treatment of CRC, which is the main limitation of such therapy, said Alberto Bardelli, PhD.
Cetuximab and panitumumab as monotherapy are effective in 15% to 20% of patients with metastatic CRC. The molecular mechanisms underlying primary resistance to anti-EGFR therapy in CRC are oncogenic activation of EGFR downstream effectors such as KRAS, BRAF, PIK3CA, and PTEN. More recently, ERBB2 amplification and MET amplification were also determined to play a role in the lack of response in this setting.
“Discovery has been associated with a lack of response rather than predictive value,” said Bardelli, associate professor, Candiolo Cancer Center, University
of Torino, Candiolo, Italy. “Nobody has determined what makes these patients sensitive to cetuximab and panitumumab.”
After the initial response to cetuximab or panitumumab, secondary resistance invariably develops. “It’s rare that somebody has a complete response with anti-EGFR therapy,” he said. “It’s fair to say that patients will relapse.”
Drivers of Secondary Resistance
The basis for secondary resistance had been poorly understood. Almost all colorectal tumors will develop with an APC/beta catenin mutation. Expression of mutant KRAS, NRAS, or BRAF mutation follows in about 50% of cases. Alternatively, colorectal tumors will evolve with a receptor tyrosine kinase alteration instead of a BRAF mutation. This alteration occurs in 20% to 30%.
The third possibility in CRC progression, in which the transition to late stage is driven by an autocrine loop, is much less frequent. This last category of colorectal tumors grows despite being sensitive to EGFR inhibitors. There is no genetic biomarker to identify this third category, said Bardelli. “Not all biomarkers will be genetically driven, and in these tumors I don’t think genetics explains why they don’t respond,” he said.
In order to understand resistance mechanisms, Bardelli and colleagues went back to the laboratory to use CRC cell lines, and also developed “xenopatients,” which are patient-derived, drug-resistant CRCs grafted and grown in mice. They currently have about 170 CRC cell lines, 12 of which were derived directly from xenopatients.
A cetuximab trial in CRC cell lines showed that 15% to 20% of cells are sensitive to cetuximab, and the ones that are not sensitive have biomarkers of resistance. “Most of the cells that respond will have a KRAS or NRAS or BRAF mutation, but in some instances we can’t explain what’s happening,” he said.
The cells that were cetuximab-sensitive were transformed through cetuximab or panitumumab treatment into cells that became resistant. “In doing so, we studied acquired resistance to EGFR blockade in CRC,” said Bardelli. “The first cell line we studied was a cell line that was highly sensitive to cetuximab, had an amplification of EGFR, and when we turned it into a resistant cell line, it lost amplification of EGFR and acquired a KRAS amplification.”
Therefore, a predominant mechanism of acquired resistance to anti-EGFR therapy is the acquisition and emergence of KRAS mutation.
“We continue to identify alleles that drive resistance,” he said. “Other pathways are involved. We recently discovered a patient that had a K57N as an acquired mechanism of resistance to EGFR therapy.” A second mechanism of acquired resistance involves mutation of the primary target.
“The 2 mechanisms are incredibly similar,” Bardelli said. “I would argue that primary and acquired resistance to EGFR are driven by the same molecular entities. Essentially, when you start treating your patients and you apply selective pressure, you simply select for alleles that will drive secondary resistance. The same alleles that drive primary resistance also drive acquired resistance. The difference is the time it took for the clones that contain these mutations to emerge and develop.”
When a biopsy is performed of CRC tissue from a patient who has acquired resistance to cetuximab, only a fraction of the cells in the biopsy carry KRAS mutations, which suggests a nongenetic mechanism of acquired resistance may also be in play.
When supernatant taken from cetuximab-resistant cells is reinserted into cells sensitive to cetuximab and panitumumab, both sensitive and resistant cells grow, which suggests that the resistance is not genetic. In this case, cetuximab-resistant cells create a permissive environment for sensitive cells. A paracrine cross talk driven by the resistant subpopulation provides protection of surrounding sensitive cells. “Only a fraction of the cells will acquire mutations, and the other cells will survive based on networks of ligands,” said Bardelli.
Liquid Biopsies: Genetic Markers in the Blood
Sampling tumor tissue has several limitations, including difficulty in obtaining the tissue and selection bias resulting from tumor heterogeneity. Biomarkers found in the blood are not novel, but the technology to measure circulating tumor DNA has evolved. Advances in the sensitivity and accuracy of DNA analysis have permitted genotyping of circulating cell-free DNA for somatic genomic alterations found in tumors. APC mutations in the blood are the “perfect specific biomarker,” he said, because they’re present early in CRC tumors, and there is little tumor heterogeneity with liquid biopsy.
“We are looking at the blood only, so we can’t say from which metastasis there is the evolution of the mutation,” said Bardelli. “But certainly we can say that the patient is having major progression associated with multiple KRAS mutations. These mutations are not stable. Some will extinguish themselves and be taken over by other mutations.”
Liquid biopsies will gain in importance in the future, Bardelli predicted. Clinical applications include monitoring response to therapy and detection of resistance mutations.
Personalized Medicine in Oncology Brings Challenges to Regulatory Agencies in Their
Appraisal Process
While personalized medicine is evolving to become an integral part of modern medicine, regulators are grappling with the assignment of value to new technologies, said Andrew Stainthorpe, PhD, who spoke on the regulatory aspects of personalized medicine in oncology.
Stainthorpe was speaking from the perspective of associate director of the National Institute for Health and Care Excellence (NICE), which is an independent organization established in the United Kingdom to provide guidance for decisions on which drugs and treatments are made available. Appraisals of new health technologies are based primarily on evaluations of efficacy and cost-effectiveness under various circumstances.
“Does the current regulatory landscape work well for manufacturers and researchers?” he asked. “And does it work well for patients, clinicians, payers, and health technology assessment [HTA] committees? In some respects it doesn’t, and that’s why perhaps you’re seeing from NICE...a number of negative decisions about the cost-effectiveness of diagnostic tests and for new technologies and the cost-effectiveness of those.”
Healthcare expenditures are outpacing the growth of the gross domestic product in most countries at a pace that is not sustainable. Key parts of regulatory agencies and HTA committees are the process by which decisions are made with budget constraints in mind.
Payers have a difficult role because they work with tightly controlled budgets, said Stainthorpe. They must be convinced of the value of medicines and new technology, especially in the context of current treatment options.
Cancer is not a uniform disease, as there are several molecular subsets of most cancers. In addition, some drugs are now effective for the treatment of several rare cancers, such as imatinib for CML with a Bcr-Abl translocation and gastrointestinal stromal tumors with KIT mutations, or trastuzumab for HER2-overexpressing breast cancer and gastric cancer. The research is leading to a plethora of different cancers, and questions arise regarding the best way to treat them “and which technologies work best to treat patients who may have more than 1 tumor type expressing a range of different mutations,” he said.
NICE typically considers a range of factors in making its guidance. It assesses the evidence put before them by manufacturers of technology. Whereas a threshold of £20,000 per quality-adjusted life year has been accepted as cost-effective, NICE may allow higher prices for innovative technologies, he said. It is in the process of establishing a formal transparent process that lays out the appraisal parameters that enter into its decisions, “so the manufacturers bringing such technology to market will know what evidence to collect and what actually makes a difference in the technology appraisal,” said Stainthorpe.
“There are technology appraisals for which a manufacturer might bring a companion diagnostic into the process,” he said. “And there’s a diagnostic appraisal program with appraisal of a new diagnostic without it being linked to a drug. Those are the 2 programs where NICE operates where a biomarker is featured.”
Often, NICE will give a qualified “yes” to a new technology, usually when in a post hoc review of the data it determines that a subset of the population benefits more than the overall population from the technology. “It may be that there’s a genetic aspect to it,” he said. “There are still situations in which there’s a good match between a companion diagnostic and its indication, but it’s not cost-effective, and NICE has said that it can’t recommend that treatment.”
The patient perspective is also considered in decisions. Evidence must show benefit of a treatment on quality of life and not only on the disease.
With personalized medicine, regulators face the challenge not only of small sample size (in some instances, N=1) but also of patients with a number of different genotypes. How that fits into decisions about therapies to make available and the cost of such medicines is
uncertain.
“It’s an exciting time, but I think we need more research and more input in a way that’s digestible for the regulators and the HTA community,” he said.

