Advocating for biomarker-driven “smart” trials, making the case for patient selection for specific drugs, and the concept of rational combinations as potential targeted therapies in order to improve the efficacy of clinical trials were topics of an insightful presentation delivered by Dr Aleksandar Sekulic on behalf of Dr Pat LoRusso at the inaugural conference of the World Cutaneous Malignancies Congress.
Sekulic reminded the audience that historically, drugs were designed, patients were treated, and individual patients had different responses in early clinical trials. The goal in the new era of personalized treatment is to shift to a different model; one that determines the molecular profile of the individual patient’s tumor and then attempts to match that profile with the available panel of targeted drugs. This approach will refocus the efforts of early clinical trials.
This pharmacodynamic approach of using predictive biomarkers leads to patients who are more likely to respond to therapy, as well as other measurements such as intermediate end point biomarkers. Sekulic cited examples of such “smart” trials: SMO in basal cell carcinoma, PARP in breast cancer, ALK and EGFR in non–small cell lung cancer, BRAF in melanoma, and c-kit in gastrointestinal stromal tumors. The objective response rates in these early trials are impressive, especially compared with the historic controls at this stage.
Sekulic also pointed out possible issues with preselecting patients: some patients who could benefit from the drug may inadvertently be excluded, and as we start breaking down common diseases into smaller subsets, there are decreasing numbers of patients on the trial, posing challenges in recruiting, biomarker cost, and quality of tissue processing.
Intratumoral Heterogeneity
Another issue Sekulic addressed is intratumoral heterogeneity. A single tumor biopsy may not represent the entire tumor population. One approach is to look at mutations and markers that can be identified with targeted assays that can be designed either to a single gene, such as BRAF, or to a panel of genes.
The next obvious step, which has been explored by several groups, is next-generation sequencing, where one can identify multiple mutations, potentially differentiate drivers versus passengers, and pursue functional studies.
Sekulic pointed out that in order to treat the entire cancer in a patient, we have to consider combination therapy. He cited a presentation by Vogelstein at the recent American Association for Cancer Research meeting describing some of the challenges that face us in this area and summarized them in 4 points:
- To better understand the pathways in human cancer cells or humanized systems
- To rationally identify the combination therapies using new screening approaches
- To develop new biologic treatments combining biologic-, immunologic-, and genomic-defined therapies
- It is not just about curing advanced cancer; prevention and early detection are a large part of the answers
Not only do we need to pick the right agents and right doses and right schedules, but also the right drug combinations. These combination strategies may be required to overcome not only cross-talk and feedback, as we have seen with recent strategies of BRAF and MEK, but also intratumoral heterogeneity.
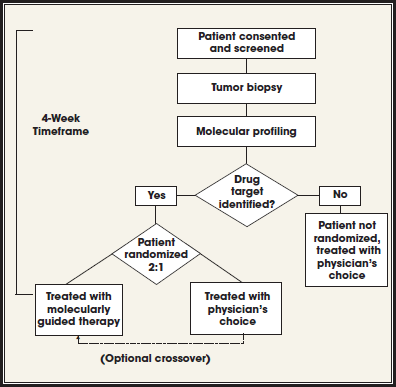
Personalized Medicine for Patients With Metastatic BRAF Wild-Type Melanoma
All these considerations are being applied to a trial designed for patients with BRAF wild-type melanoma led by Jeff Trent and Pat LoRusso. In the proposed flowchart of the trial, the tissue is obtained, genome is profiled, data are processed, knowledge mining to identify pathways affected is performed, and clear validation assays are conducted, after which the tumor board meets and the therapy is selected. In conjunction, a set of translational experiments in mouse explants and cell lines will be carried out to test some of the predictions of responses.